Virus Host Cell Genetic Material Transport
William E. Schiesser
* Affiliatelinks/Werbelinks
Links auf reinlesen.de sind sogenannte Affiliate-Links. Wenn du auf so einen Affiliate-Link klickst und über diesen Link einkaufst, bekommt reinlesen.de von dem betreffenden Online-Shop oder Anbieter eine Provision. Für dich verändert sich der Preis nicht.
Springer International Publishing 
Naturwissenschaften, Medizin, Informatik, Technik / Biologie
Beschreibung
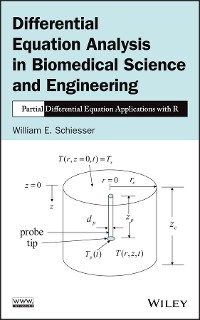
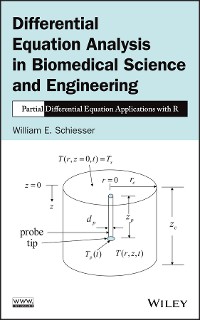
The reproduction and spread of a virus during an epidemic proceeds when the virus attaches to a host cell and viral genetic material (VGM) (protein, DNA, RNA) enters the cell, then replicates, and perhaps mutates, in the cell. The movement of the VGM across the host cell outer membrane and within the host cell is a spatiotemporal dynamic process that is modeled in this book as a system of ordinary and partial differential equations (ODE/PDEs).
The movement of the virus proteins through the cell membrane is modeled as a diffusion process expressed by the diffusion PDE (Fick’s second law). Within the cell, the time variation of the VGM is modeled as ODEs. The evolution of the dependent variables is computed by the numerical integration of the ODE/PDEs starting from zero initial conditions (ICs). The departure of the dependent variables from zero is in response to the virus protein concentration at the outer membrane surface (the point at which the virus binds to the host cell).
The numerical integration of the ODE/PDEs is performed with routines coded (programmed) in R, a quality, open-source scientific computing system that is readily available from the Internet. Formal mathematics is minimized, e.g., no theorems and proofs. Rather, the presentation is through detailed examples that the reader/researcher/analyst can execute on modest computers. The ODE/PDE dependent variables are displayed graphically with basic R plotting utilities.
The R routines are available from a download link so that the example models can be executed without having to first study numerical methods and computer coding. The routines can then be applied to variations and extensions of the ODE/PDE model, such as changes in the parameters and the form of the model equations.
Kundenbewertungen
Host Cell Proteins, genetic material, cellular membrane, R routines, RNA, viral genetic material, ODE/MOL routine, ODE/PDE model, DNA, Postulated vaccine, single protein model, virus replication, numerical integration, mutate, Postulated therapeutic, virus protein